Home / Challenge areas / CanCOGeN / VirusSeq
VirusSeq

CanCOGeN’s VirusSeq initiative is sequencing up to 150,000 viral samples from people testing positive for COVID-19 with more coordination of viral sequencing and associated clinical/epidemiological data collection between provinces and internationally.
This effort is enabling Canada to track how the SARS-CoV-2 virus that causes COVID-19 is changing and spreading, providing key data on virus transmission trends and changes in the virus that may impact viral detection or the effectiveness of treatments or vaccines.
As SARS-CoV-2 spreads between individuals, the virus continues to mutate. Tracking these changes is possible through genome sequencing of the virus. These changes can then be used to generate a “family tree” for this virus and its “descendants,” aiding researchers in the linking of related cases. Changes in the virus can also change disease severity or its ability to spread. Viral genome sequence data has therefore become an essential tool for public health officials to better track how the virus is both changing and spreading.
The virus timeline
November 2019
The human coronavirus SARS-CoV-2, the virus responsible for the COVID-19 outbreak, first emerged in late November 2019 and within four months reached pandemic status, spreading to at least 177 countries and resulting in one of the largest shutdowns of human activities in modern history.
January 2020
Critical to global efforts to control COVID-19 and its impacts has been the rapid release of the first genomic sequence on Jan 10, 2020. This not only enabled initiation of vaccine and antiviral therapeutic development, but also enabled key molecular diagnostic tests to be designed and shared within a month of the disease being reported to the WHO. These diagnostic tests, however, only have the resolution to confirm whether a person has COVID-19 or not. Genomic sequences additionally identify how related cases are to each other, so they can be used to track where the virus came from and further identify how it is spreading nationally and internationally. Canada.
Genome Canada leads in proposing new national viral genomics network.
February 2020 to present
Genomic sequences, rapidly shared with contextual data, have been key to confirm the source of cross-border introductions of the virus, and the identification of community transmission — informing policies for outbreak control. Continued genomic sequencing efforts are critical for modeling of the local incursion and spread of the virus. As the number of community-transmitted COVID-19 cases increases, epidemiological investigation and tracing will increasingly rely on genomic data as evidence to detect clusters. Continued genome-based surveillance after the number of cases reduces is also essential to identify and control the sources of clusters that risk a second epidemic wave.
Looking ahead
Genomic sequence changes associated with severity of disease or eventually drug resistance can also be tracked and potentially inform patient management as the virus evolves.
Click here to learn more about the governance of CanCOGeN VirusSeq.
Why Sequence this virus?
Identify and track transmission trends at the regional, provincial, national and international scales
Aid detection of new clusters of cases/outbreaks
Discover evolving viral characteristics that might impact
• Detection methods (PCR, serology)
• Clinical outcomes (strain severity) and transmission
• Effectiveness of healthcare measures, treatments and vaccines

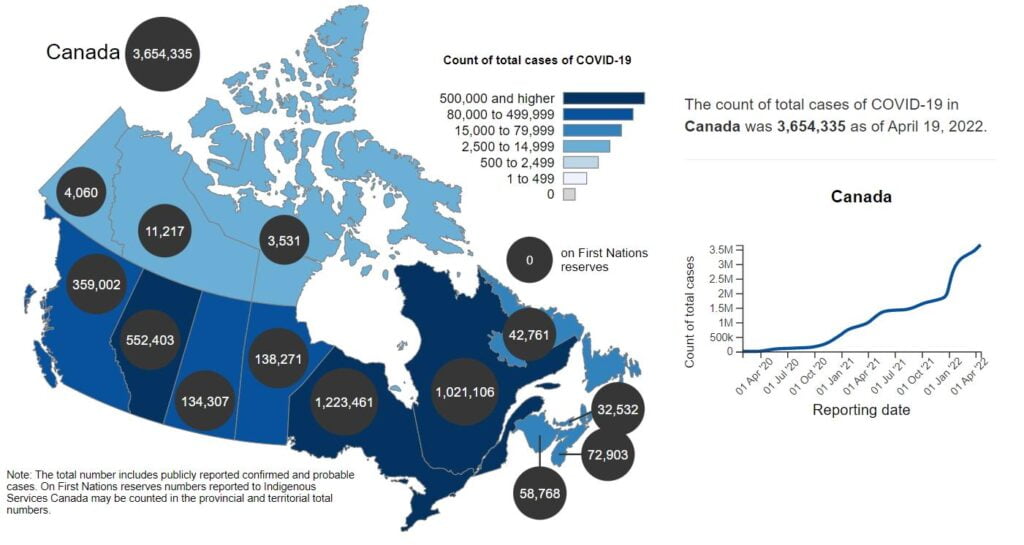
Coronavirus disease (COVID-19): Outbreak update
See the current situation with total cases, active cases, recovered cases, tests performed or deaths in Canada over time.
Resources
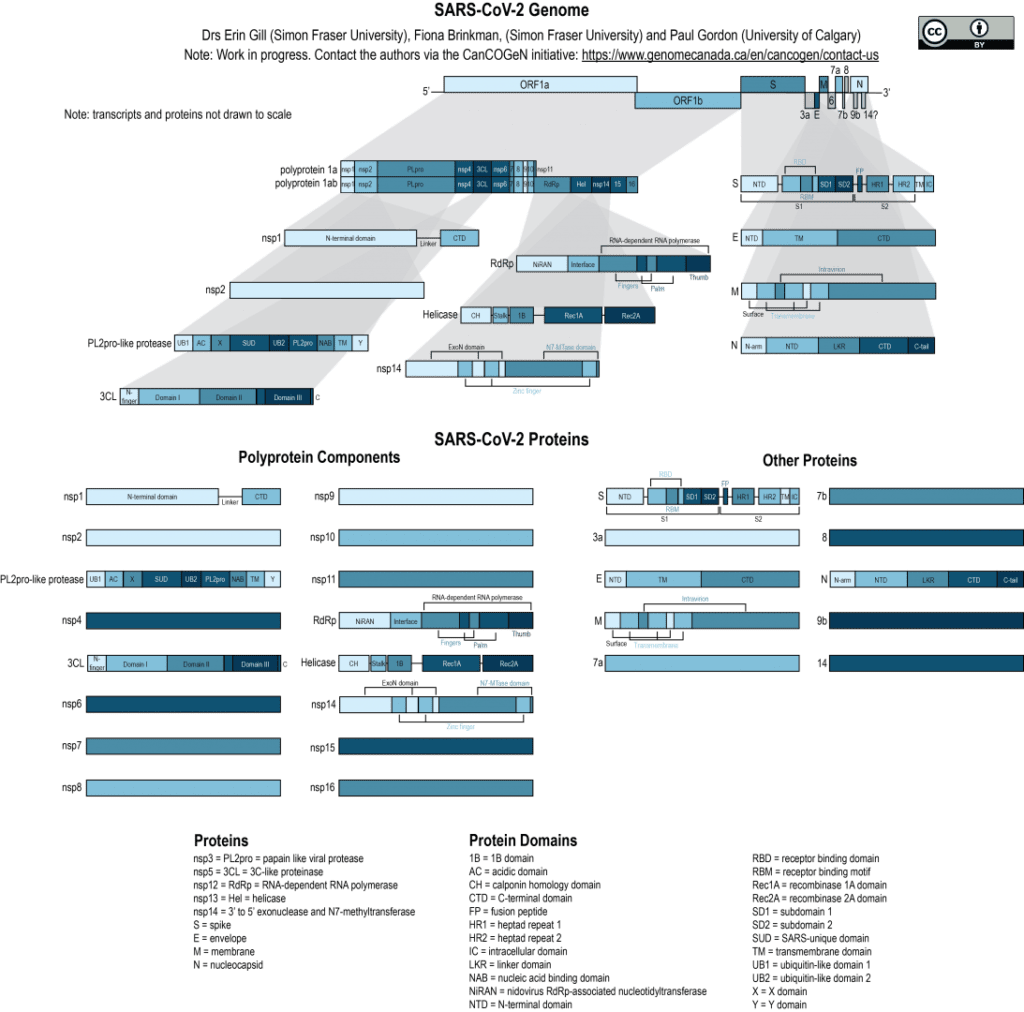
Schematic of the SARS-CoV-2 genome, illustrating the complexity of the proteins that it encodes. The genome contains two open reading frames (ORF1a and ORF1b) that encode polyproteins 1a and 1b. These polyproteins are cleaved into their individual protein components following translation. The genome also encodes other proteins, including the structural proteins (most notably the Spike/S protein/gene), that are translated independently. Each protein encoded by the SARS-CoV-2 genome is displayed individually, and different functional/structural domains are annotated within selected proteins.
Resources
Schematic of the SARS-CoV-2 genome, illustrating the complexity of the proteins that it encodes. The genome contains two open reading frames (ORF1a and ORF1b) that encode polyproteins 1a and 1b. These polyproteins are cleaved into their individual protein components following translation. The genome also encodes other proteins, including the structural proteins (most notably the Spike/S protein/gene), that are translated independently. Each protein encoded by the SARS-CoV-2 genome is displayed individually, and different functional/structural domains are annotated within selected proteins.